

 Mutation
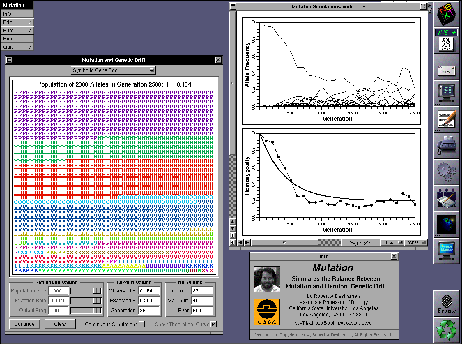
MutationMutation was created for an introductory college course in population genetics. It simulates stochastic fluctuations in the genetic composition of a population due to the balance between random genetic drift and mutation. Population size can vary from one hundred to one thousand diploid individuals and mutation rates between 0.001 and 0.0001 per allele per generation can be selected. Students can switch among three different views: allele frequencies versus time, mean homozygosity versus time, or a symbolic representation of the gene pool. In the latter view, alleles are represented by different letters and different colors. The application shows running values for the observed and expected homozygosity, the current number of alleles, the maximum number of alleles that ever existed in the population, and the total number of alleles produced by mutation. Graphs can printed or copied and pasted into other applications. A screen image of a Mutation session appears below. Mutation was written by Dr. Robert Desharnais.
Click to download full resolution image (99.9 KB.)
 Return to the Application Catalog
Return to the Application Catalog